Distribution of Bemisia tabaci in different agro-ecological regions in Uganda and the threat of vector-borne pandemics into new cassava growing areas
- 1Natural Resources Institute, University of Greenwich, Kent, United Kingdom
- 2Root Crops Programme, National Crops Resource Research Institute, Kampala, Uganda
- 3Department of Entomology, The Hebrew University of Jerusalem, Rehovot, Israel
Previous studies in sub-Saharan Africa have showed the spread of cassava mosaic disease (CMD) and cassava brown streak disease (CBSD) pandemics into different cassava growing regions by high Bemisia tabaci populations. Studies did indicate that there were stark differences in some whitefly species, yet they have not looked extensively across agroecologies. Members of B. tabaci species complex termed sub-Saharan Africa 1 (SSA1) and SSA2 have been linked to the spread of CMD and CBSD viruses. During the period of a severe CMD pandemic in the 1990s, SSA2 was the most predominant until the resurgence of SSA1, particularly SSA1-subgroup1 (SSA1-SG1) from the early 2000s to date. Cassava being a drought resilient crop has become an important food security crop and has been introduced into new areas and regions. Considering the role B. tabaci in the spread of cassava virus pandemics into neighboring regions, we investigated the genetic diversity and distribution of B. tabaci in nine different agro-ecological regions of Uganda in 2017. Adult whiteflies were collected from cassava and 33 other host plants from cassava-growing areas, those with limited cassava and areas with no cassava, where it is being introduced as a food security crop. The partial sequences of the mitochondrial cytochrome oxidase 1 (mtCO1) gene (657 bp) were used to determine the phylogenetic relationships between the sampled B. tabaci. Cassava B. tabaci SSA1 (-SG1, -SG2, -Hoslundia (previously called SSA1-SG1/2), -SG3), SSA2 and SSA3; non-cassava B. tabaci SSA6, SSA10, SSA11, SSA12, SSA13, MED-ASL, MED-Q1, MEAM1, Indian Ocean; and other Bemisia species, Bemisia afer and Bemisia Uganda1 were identified in the study. SSA3, one of the key B. tabaci species that occurs on cassava in West Africa, was identified for the first time in Uganda. The SSA1-SG1 was widely distributed, predominated on cassava and was found on 17 other host-plants. The ability of SSA1-SG1 to exist in environments with limited or no cassava growing poses the risk of continued spread of virus pandemics. Therefore, measures must be put in place to prevent the introduction of diseased materials into new areas, since the vectors exist.
Introduction
Cassava (Manihot esculenta) is one of the most important staple food crops for over 800 million people and source of income in sub-Saharan Africa (Burns et al., 2010; FAO, 2019). Its ability to grow in marginal areas where other crops cannot survive and to store roots for lengthy periods in the ground allowing flexible harvesting over long periods, has made it an important food security crop (El-sharkawy, 2007; Rey and Vanderschuren, 2017). Cassava production is mainly constrained by two devastating diseases, cassava mosaic disease (CMD) and cassava brown streak disease (CBSD). Both are caused by viruses transmitted by B. tabaci (Maruthi et al., 2002, 2005; Legg et al., 2014).
CMD is caused by cassava mosaic geminiviruses (CMGs) belonging to the genus Begomovirus and family Geminiviridae (Bull et al., 2006; Alabi et al., 2011). CMD symptoms are characterized by mosaic patterns on the leaves, leaf distortion and reduced leaf size, which reduces the photosynthetic area and hence plant growth and yields. Plants that are severely affected become highly stunted and produce no tubers (Alabi et al., 2011). CMD is wide spread in sub-Saharan Africa (Rey and Vanderschuren, 2017) and a total yield loss of US$1.9–2.7 billion annually has been reported in all affected countries (Scholthof et al., 2011).
CBSD is caused by cassava brown streak viruses (CBSVs) that belong to genus Ipomovirus and family Potyviridae (Monger et al., 2001; Mbanzibwa et al., 2009; Winter et al., 2010; Ndunguru et al., 2015). CBSD symptoms include yellow blotchy chlorosis in the primary and secondary leaf veins occurring on mature leaves, brown necrotic streaks on the green portions of the stems, root constriction and yellow or brown corky root necrosis that renders the tubers unsuitable for consumption (Alicai et al., 2007). CBSD has been reported in eastern Africa and around the Great Lakes region (Rey and Vanderschuren, 2017), with an estimated yield loss of US$750 million (Alicai et al., 2019).
As a vegetatively propagated crop, these viruses are passed from one generation to the next and easily accumulate within a plant among the susceptible genotypes. This allows recombination thus causing rapid spread of diseases and virus variants that in some cases have resulted in severe pandemics, for example, the cassava mosaic pandemic of the late 1990s (Zhou et al., 1997). On the other hand, reports indicate that the genome of cassava brown streak virus is rapidly evolving (Alicai et al., 2016), with more genetic variations in cassava brown streak viruses (Ndunguru et al., 2015). Management of these viral diseases has focussed on continued development and deployment of CMD resistant and CBSD tolerant cassava varieties (Legg et al., 2011; Kawuki et al., 2016). Efforts have been put in place especially by the government of Uganda to ensure that its National Agricultural Research Systems distribute virus-free materials to farmers. There are also ongoing developments to produce cassava varieties with both virus and whitefly resistance.
Bemisia tabaci is a group of at least 40 putative cryptic (morphologically indistinguishable) member species (Dinsdale et al., 2010; Liu et al., 2012; Legg et al., 2014; Mugerwa et al., 2018, 2021a). B. tabaci cause damage through: (i) direct feeding that causes stunted growth and plant necrosis especially on young plants, (ii) secretion of honeydew which supports saprophytic fungi (sooty mold) that reduce photosynthesis and fruit quality (Byrne and Bellows, 1991; Brown et al., 1995; Oliveira et al., 2001), and (iii) transmission of >400 virus species (https://ictvonline.org/taxonomy), the majority of which belong to the genus Begomovirus (Brown et al., 2015; Zerbini et al., 2017). Other economically important viruses spread by B. tabaci consist of torradoviruses, criniviruses and ipomoviruses (Jones, 2003; Winter et al., 2010; Gilbertson et al., 2015; Lecoq et al., 2016).
The partial 3' mitochondrial cytochrome oxidase 1 (mtCO1) sequences have been used widely to delimit cryptic species within the B. tabaci complex, although it is increasingly becoming recognized that an integrative approach that combines molecular marker information and biological studies is required to discover the full species diversity within B. tabaci (Vyskočilová et al., 2018; Mugerwa et al., 2020). In Africa, putative species of B. tabaci called sub-Saharan Africa (SSA) [SSA1 to SSA7 and SSA9 to SSA16], Mediterranean (MED), East Africa 1 (EA1), Indian Ocean (IO) and Middle East-Asia Minor1 (MEAM1), colonize important agricultural crops including Manihot esculenta (cassava), and Ipomoea batatas (sweetpotato), horticultural crops such as Cucurbita spp., Phaseolus vulgaris (beans), Abelmoschus esculentus (okra) as well as various weed species (Sseruwagi et al., 2005; Esterhuizen et al., 2013; Legg et al., 2014; Mugerwa et al., 2018, 2021a; Vyskočilová et al., 2019). SSA1 and its different sub-groups (1 to 5 and 1/2), SSA2, SSA3, SSA4 and SSA5 utilize cassava as a host-plant for development (Berry et al., 2004; Esterhuizen et al., 2013; Legg et al., 2014; Manani et al., 2017; Wosula et al., 2017).
The severe CMD pandemic that caused tremendous yield losses in Uganda in the 1990s (Otim-Nape et al., 1994) and CBSD pandemic in the 2000s (Alicai et al., 2007) have since spread to other east and central African countries. The spread of these pandemics to neighboring regions has been associated with high populations of B. tabaci known as SSA2 (Legg et al., 2002), which was later replaced by SSA1, largely SSA1-SG1 from the 2000s to date (Mugerwa et al., 2012; Legg et al., 2014; Ally et al., 2019). It was reported that the CMD pandemic in western Kenya in 2001 was caused by SSA2 that migrated from eastern Uganda, where the CMD pandemic is reported to have started in the late 1980s (Legg, 1994; Legg et al., 2014; Manani et al., 2017). The increase in CMD in north-eastern Rwanda and north-eastern Burundi in 2000 and 2003, respectively, was linked to the abundance of SSA1-SG1 that was migrating from Uganda (Legg et al., 2014). The high populations of SSA1-SG1 in Central African Republic (CAR) were also associated with the spread of CMD through Democratic Republic of Congo (DRC) that had occurred from Uganda (Tocko-Marabena et al., 2017). The severe CMD pandemic in Tanzania was attributed to the high populations of SSA1-SG1 that spread it from Uganda through the neighboring districts of Misenyi and Geita (Tajebe et al., 2015).
A number of propositions to the high whitefly populations have been put forward and these include: (i) virus-vector-host interactions that increased the fecundity of whitefly populations on cassava, leading to vectors that spread the disease (Colvin et al., 2004), (ii) CMD-resistant but highly susceptible whitefly cassava varieties that altered the ecological conditions to favor the creation of high whitefly populations (Omongo et al., 2012), and (iii) the arrival of an invader vector population (now known as SSA2) that was better adapted to cassava, more highly fecund and more efficient at transmitting the East African cassava mosaic virus-Uganda variant (EACMV-UG) than the indigenous SSA1 species (Legg et al., 2002). In addition, SSA1-SG1 has been reported to occur on more than 30 host-plant species compared to other mtCO1 subgroups in the SSA1 species (Tocko-Marabena et al., 2017; Mugerwa et al., 2021a), suggesting the role of other host-plants in its ecology and population dynamics. A wide host-range may enable such insect pests to become highly abundant, even in the absence of their most preferred host-plant species (Joyce, 1983; Butler and Henneberry, 1985). Previous studies in east and central Africa that identified B. tabaci namely SSA2 and SSA1-SG1, as responsible for the CMD pandemic (Legg et al., 2002, 2014; Mugerwa et al., 2012) were carried out mainly in cassava growing areas. The area under cassava cultivation is expanding to include areas not previously known to grow cassava like Karamoja sub-region and the highland ranges. This is because cassava has been identified as a food security crop that can withstand the effects of climate change (Jarvis et al., 2012). Although surveys on the B. tabaci associated with the pandemics in East Africa have indicated spread of high whitefly populations migrating into new regions, recent studies have shown that East Africa is the center of origin of B. tabaci (Mugerwa et al., 2018). The cassava viruses are also endemic to Africa (Tiendrébéogo et al., 2012; Tomlinson et al., 2017). It is likely that these whitefly species could exist in non-cassava growing areas on other host-plants. This study sampled over a wide area, across agro-ecological regions in Uganda to generate a greater understanding of the diversity of whiteflies present on cassava and alternative hosts in the different agro-ecological regions. This study revealed the presence a cassava whitefly species (SSA3) not previously known to exist in Uganda. Such knowledge is vital when developing and deploying appropriate integrated management strategies aimed at controlling emerging outbreaks of whitefly and whitefly-transmitted diseases. The agro-ecological zones were classified based on differences in farming systems, edaphic factors, climatic conditions, altitude and major vegetation cover (Kraybill and Kidoido, 2009).
Materials and methods
Study area
The study was conducted in nine agro-ecological regions in Uganda in May and August of 2017 (Figure 1). The study areas included cassava growing areas, those with limited cassava growing, and areas with no cassava. Cassava growing areas included: the north western savanna grassland (Lira district), north eastern savanna grassland (Otuke district) and para savannas (Kiryandongo district) are characterized by monomodal rainfall, with an annual average rainfall of 1,340, 1,197, and 1,259 mm, respectively. The regions have medium to high productive soils. The main crops grown are pulses, groundnuts, sorghum, millet, okra, sesame and cassava. Farming is mainly subsistence with a few commercial farms. Kyoga plains (Kamuli, Iganga, Budaka, Mbale, Soroti and Serere districts), Lake Victoria crescent (Mukono, Wakiso and Jinja districts) and western savanna grasslands (western Masindi, Kyenjojo, Kabarole, southern Kasese and Kamwenge districts) are characterized by bimodal rainfall, with annual averages of 1,215, 1,750, and 1,270 mm, respectively. The soils in the Kyoga plains are poor/medium productive, whereas those in the Lake Victoria crescent and western savanna grasslands are medium/high productive. Crops grown in the three regions include maize, pulses, sweetpotato, groundnuts, robusta coffee, horticultural crops, bananas and cassava. Kyoga plains and western savanna grasslands grow millet and each region exclusively grows cotton and tea, respectively. Farming in the Kyoga plains is subsistence with some commercial farming, while the Lake Victoria crescent and western savanna grasslands have both subsistence and commercial farming (Kraybill and Kidoido, 2009). The altitude in the sampled cassava growing areas ranges between 900 to 1,570 m above sea level (a.s.l) and annual average temperature ranging from 20.3 to 24.4°C (Climate-Data.org).
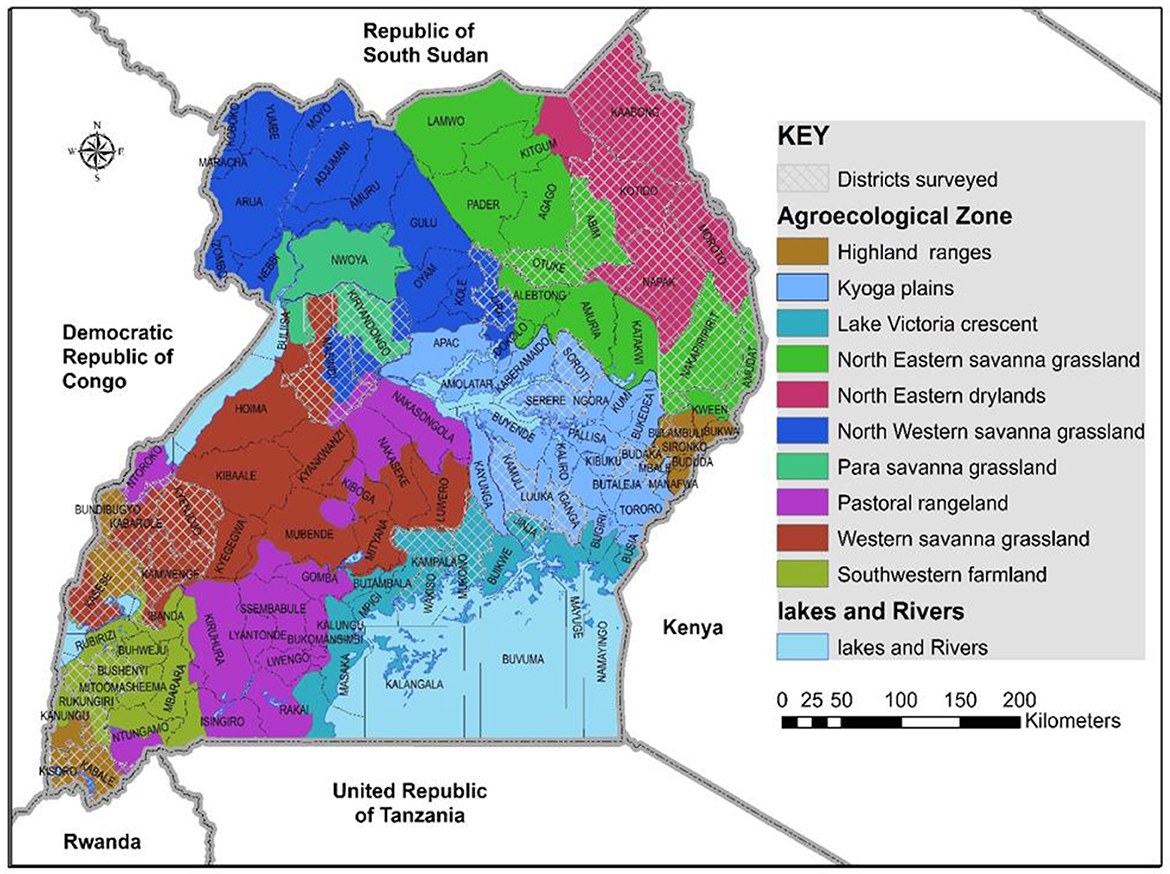
Figure 1. Agro-ecological areas in Uganda where whiteflies were collected during May/August 2017 are shown by the white-mesh covered areas.
Areas with limited cassava growing were: Karamoja's north eastern savanna grassland (Abim, Nakapiripirit and Amudat districts) is characterized with monomodal rainfall with an annual average ranging from 300 to 1,000 mm (USAID, 2017a) and annual average temperature between 20 and 24°C (Climate-Data.org). The altitude in the sampled areas ranges from 1100 to 1490 m a.s.l. The soils are poor/medium productive and farming is predominantly subsistence. Crops grown include sesame, sunflower, groundnuts, sorghum. South western farmland (Rukungiri and Bushenyi districts) and highland ranges (Bulambuli, Bundibugyo, northern Kasese, Kisoro and Kabale districts) are characterized by bimodal rainfall with annual averages of 1,170 and 1,400 mm, respectively (Kraybill and Kidoido, 2009). The altitude in the sampled south western farmland areas ranges between 1, 590 and 1630 m a.s.l and annual average temperature ranging from 19.5 to 21°C (Climate-Data.org). The altitude in the sampled highland ranges is between 850 and 2010 m a.s.l and annual average temperature ranging from 16.8 to 21.2°C. Crops grown are bananas, beans, sweetpotato and maize. South western farmland also grows robusta coffee and millet, whereas the highland ranges grow Arabica coffee, potatoes and peas.
Areas with no cassava growing included: the north eastern drylands (Kaabong, Kotido, Moroto, and Napak districts) are characterized by monomodal rainfall with an annual average of 745 mm. The soils are poor/medium productive. Farming is largely subsistence with pastoral activities, Crops grown include gum arabica, sesame, sorghum and sunflower (Kraybill and Kidoido, 2009). The annual average temperature is 20–24°C and altitude between 1,000 and 1,500 m a.s.l.
Whitefly collection
Adult whiteflies were collected from cultivated and non-cultivated plants in the field at the end and beginning of the rains (May/August, 2017) in Uganda. Sampled sites were selected based on observation of whiteflies on plants, sampling along main and rural roads separated by an interval of 10–20 km as described by Mugerwa et al. (2018). For sites with cassava, whiteflies were collected from cassava and other host plants found within or adjacent to cassava fields. Plants adjacent to cassava fields were defined as those positioned in a margin of 0–25 m away from the edges of the nearest cassava field. Adult whiteflies were collected using an aspirator and preserved in 90% ethanol. Whiteflies collected from the same host-plant in a sampled site were stored in the same tube, but those from different host-plants were stored in separate tubes. A total of 289 samples was collected. Geo-coordinates (latitude and longitude) were recorded for each collection site using a Geographical Positioning System (GPS, Garmin eTrex Vista Cx) and other meta-data including the village, sub-county and district names. The geo-coordinates for each collection site were used to generate a map using the ArcMap software program version 10.6.1 (http://www.arcgis.com/features/).
Whitefly DNA extraction
Genomic DNA was extracted separately from two individual whiteflies randomly selected from each sample (Eppendorf tube), using a Chelex method developed by White et al. (2009). Each whitefly in 75 μL of 10% Chelex® (Sigma Aldrich, St Louis, USA) containing 6 μL of 10 mg/mL proteinase K was crushed in a 1.5 mL Eppendorf tube with zirconium oxide beads using a bullet blender homogeniser (Next Advance, Averill Park, NY). The Eppendorf tubes with the extract were centrifuged for 1 min at 15,871 g. The extract was then incubated for 1 h at 37°C, then for 10 min at 96°C to denature the enzyme. The DNA was extracted from 578 whiteflies and extracts were stored at −20°C until use.
PCR amplification of mtCO1 DNA, purification and sequencing
The 3' region of the mtCO1 gene was amplified using a forward primer, 2195Bt (TGRTTTTTTGGTCATCCRGAAGT) and reverse primer C012/Bt-sh2 (TTTACTGCACTTTCTGCC) with an expected amplicon size of 867 bp (Mugerwa et al., 2018). The total reaction mixture of 30 μL contained 2 μL of the DNA extract, 0.2 μM of each primer pair, 6 μL of 5X MyTaq Buffer HS (Bioline), 1.0 U of MyTaq Polymerase (Bioline) and molecular grade water (Sigma-Aldrich, UK). PCR cycling was performed in a Veriti 96 Well Thermal cycler (Applied Biosystems, UK) programmed as follows: Initial denaturation at 95°C/3 min, 10 cycles of 95°C/30 s, annealing at 45°C/30 s and 72°C/1 min; 20 cycles of 95°C/30 s, annealing at 50°C/30 s and 72°C/1 min and final cycle at 72°C/10 min. Electrophoresis of PCR products was done on 1% (w/v) agarose gels in 0.5 X Tris/borate/EDTA buffer, stained with RedSafe™ (iNtRON Biotechnology, Korea). PCR products on the agarose gel were visualized under UV light at 302 nm on a trans-illuminator GBOX, CHEMI 16 (Syngene, UK), and those with the expected size were purified using the Exonuclease 1 and Antarctic Phosphatase (Exo1-AP) treatment by Werle et al. (1994) with modifications in concentration and incubation conditions. Werle et al. (1994) used 2 μL of PCR product mixed with 1 U Exo1 and 1 U AP and incubated for 1 h at 37°C. The enzymes were inactivated for 15 min at 72°C. In this study, PCR products (20 μL) were mixed with 1 U Exo1 and 1 U AP (New England Biolabs, UK) and incubated for 20 min at 37°C. The enzymes were inactivated for 10 min at 80°C. Purified PCR products were sent for Sanger sequencing in both directions at GATC Biotech (Germany).
Phylogenetic analysis of partial mtCO1 sequences
The partial mtCO1 Sanger sequences generated in this study were first screened using quality checks in Geneious version 10.2.3. First, a consensus sequence was generated using the reverse and forward sequences for each individual whitefly (Kearse et al., 2012). The new consensus sequences of this study were aligned with high-throughput sequencing (HTS)-derived full mitogenome sequences downloaded directly from GenBank, using MAFFT (Katoh and Standley, 2013). All Sanger sequences which contained indels were eliminated and not considered for further analysis. The remaining Sanger sequences together with HTS-sequences were then trimmed to 657 bp and translated to amino acid residues from appropriate codon positions using the invertebrate mitochondrial DNA genetic codes to: (i) identify potential premature stop codons and (ii) enable amino acid residue alignment against the HTS reference CO1 amino acid data set. Sanger sequences which had premature stop codons and amino acid substitutions in highly conserved regions as identified within the trimmed HTS reference CO1 gene set were eliminated. Then 66 unique haplotypes were extracted from the remaining sequences (n = 328) and aligned with equivalent reference whitefly sequences downloaded directly from the GenBank in accordance with Kunz et al. (2019). MEGA-X was used to determine the best substitution model. Phylogenetic analysis was done using BEAST2 that employs Markov Chain Monte Carlo (MCMC) sampling to approximate posterior probabilities of phylogenies (Peter, 1995). It also utilizes the BEAGLE library, an application programming interface and library of high-performance statistical phylogenetic inference (Ayres et al., 2012). BEAUti version 2.5.1 was used to generate the xml file for analysis in BEAST version 2.5.1. Tree prior was set to “Yule”, the clock model was set to strict clock and the birth rate was uniform. The site model was specified as TN93, base frequencies were estimated and Gamma plus invariant sites was the site heterogeneity model. The MCMC were run for 10 million generations and sampled every 1,000th generation (the first 10% of trees were discarded as burn-in). TreeAnnotator version 2.5.1 was used to generate a final tree, which was viewed in FigTree version 1.4.3 to visualize the similarities between reference sequences and haplotype sequences from our study. Bemisia afer (accession no. KF734668) and T. vaporariorum (accession no. AY521265) were used as outgroups for the entire B. tabaci complex. The 66 unique haplotype sequences were deposited in the GenBank sequence database (accession numbers MN271483–MN271548).
Results
Sampling and phylogenetic analysis
The DNA was extracted from two individual whiteflies and sequenced per specific host-plant sampled in a given location. A total of 328 mtCO1 high-quality sequences from individual whiteflies were analyzed after removal of 148 sequences that contained errors/pseudogenes as described by Vyskočilová et al. (2018). The identities of the individual whiteflies were determined based on their phylogenetic placement and sequence identity with already defined species (Figure 2), using the 3.5% divergence threshold criterion in the nucleotide sequence (Dinsdale et al., 2010). Thirteen genetically distinct groups were identified in the study (Figure 3), of which six; SSA1, SSA2, and SSA3, MED-ASL, IO and B. afer were collected from cassava (Table 1). The SSA1 consisted of four sub-groups namely SSA1-SG1, SSA1-SG2, SSA1-SG3, and SSA1-IntSG1/2, which we shall refer to herein this study as the SSA1-Hoslundia because the name “Intermediate” is a misnomer. It is a sub-group of SSA1 in its own right with biological traits that separate it from the other groups, e.g., fourth instar nymphs were first identified on Hoslundia opposita and no nymphs have ever been detected on cassava in the field (Namuddu, 2019). SSA1-SG3 was not detected on cassava but on beans (Table 1). Of all the whitefly samples collected from cassava (111), SSA1-SG1 was the most predominant constituting 75.7% (84/111). The SSA1-SG2 were 11.7% (13/111), 0.9% (1/111) was SSA1-Hoslundia, 2.7% (3/111) were SSA2, 0.9% (1/111) was SSA3, 2.7% (3/111) were MED-ASL, 0.9% (1/111) was IO and 4.5% (5/111) were B. afer (Table 1).
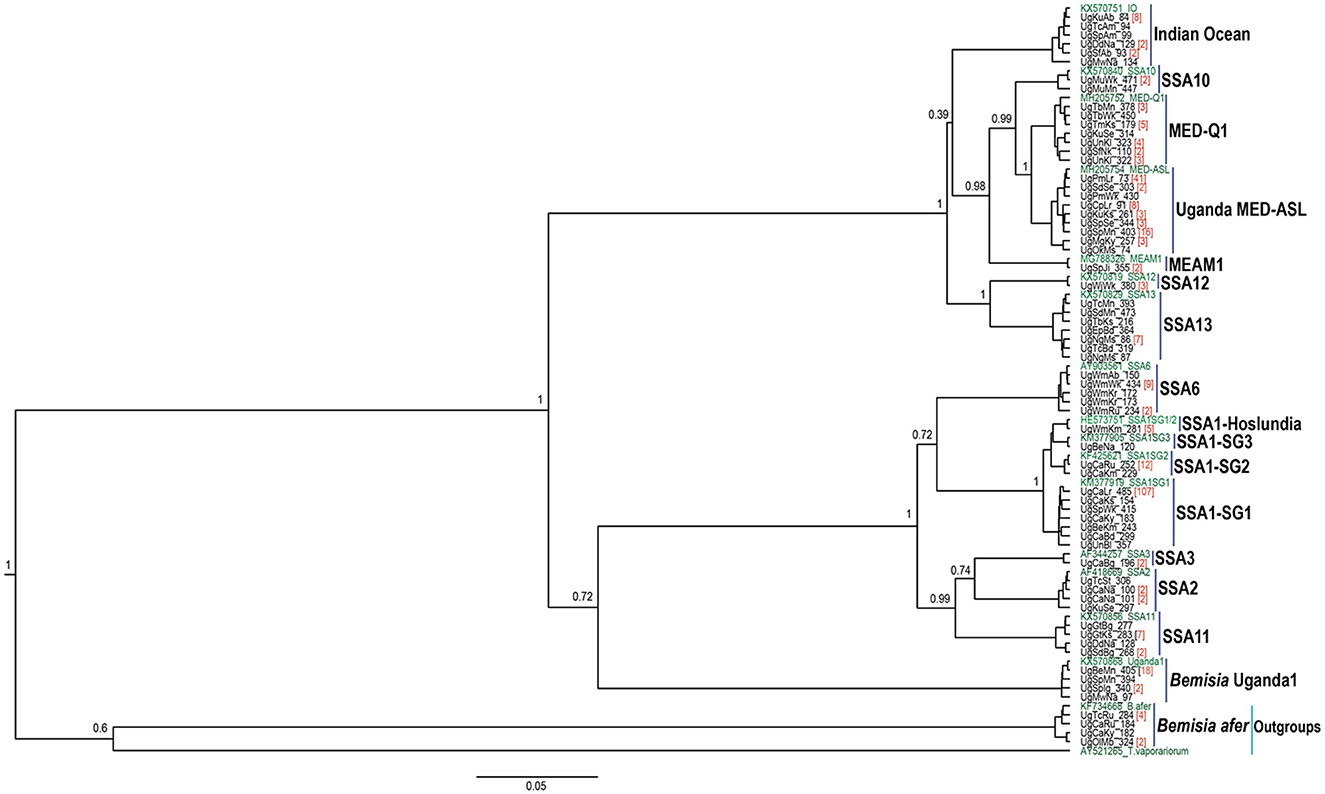
Figure 2. BEAST2 phylogenetic tree based on partial mtCO1 sequences (657 bp) of adult B. tabaci species and other whitefly species collected from cassava and common host-plants during May/August 2017 in Uganda. Numbers noted at the nodes are posterior probabilities. Numbers in red besides a sample sequence are the number of individuals with that particular haplotype and in green are reference sequences.
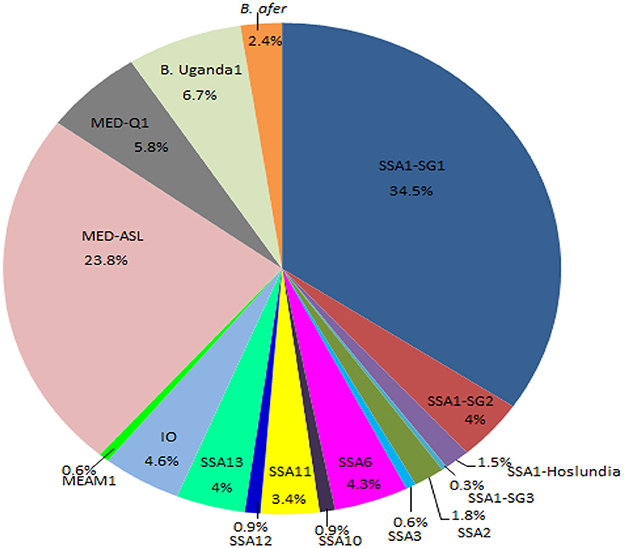
Figure 3. Percentage composition of all the whitefly species collected from the different agro-ecological regions in Uganda during May/August 2017. SSA, sub-Saharan Africa; SG, sub-group; MED, Mediterranean; ASL, African silver leafing; IO, Indian Ocean; B. afer, Bemisia afer; MEAM1, Middle East-Asia Minor1 and B. Uganda1, Bemisia Uganda1.

Table 1. Numbers of adult B. tabaci species and other whitefly species collected from cultivated and non-cultivated host-plants during May/August 2017 in Uganda.
Host-plant range of whiteflies identified in the different agro-ecologies in Uganda
Whiteflies were collected from nine agro-ecological zones in 30 districts (Figure 1) and 34 host-plant species belonging to 17 plant families (Table 1). SSA1-SG1 predominated on cassava, whereas MED-ASL was the most prevalent on non-cassava host-plants (Table 1). SSA1-SG1 constituted 34.5% (113/328), while MED-ASL consisted of 23.8% (78/328) of the total number of whiteflies typed from the different host-plants in Uganda (Figure 3). SSA1-SG1 was also found on 17 host-plant species (Table 1). The other three subgroups of SSA1 species identified in this study; SSA1-SG2, SSA1-SG3 and SSA1-Hoslundia, consisted of 4%, 0.3% and 1.5% of total typed whiteflies, respectively (Figure 3). SSA1-SG2 and SSA1-SG3 occurred on cassava and P. vulgaris, respectively (Table 1), whereas SSA1-Hoslundia was found on cassava, L. camara and O. gratissimum (Table 1). MED-ASL was found on 21 plant species and predominated on majority of them except O. gratissimum, N. tabacum, L. camara, C. benghalensis and A. conyzoids (Table 1).
Other B. tabaci species identified in the study include: SSA2, SSA3, SSA6, SSA10, SSA11, SSA12, SSA13, IO, MEAM1, MED-Q1 and these comprised of 1.8, 0.6, 4.3, 0.9, 3.4, 0.9, 4, 4.6, 0.6, 5.8%, respectively, of the total whiteflies typed (Figure 3). Other whitefly species were B. Uganda1 and B. afer and consisted of 6.7 and 2.4%, respectively (Figure 3). Whitefly species SSA2, SSA3, IO and B. afer occurred on cassava. SSA2 was also found on E. heterophylla, L. camara and H. opposita, whereas SSA3 occurred on P. vulgaris. B. afer occurred on L. camara, O. latifolia and an unknown weed species (Table 1). SSA6 was found only on O. gratissimum (Table 1). MED-Q1 was found mostly on N. tabacum. The IO and SSA13 similarly occurred on L. camara. SSA11 and SSA12 were equally found on C. benghalensis (Table 1). SSA10 was found only on V. amygdalina. B. Uganda1 commonly occurred on I. batatas (Table 1).
Distribution of whiteflies in the different agro-ecologies in Uganda
Figure 4 details the agro-ecological distributions of the different B. tabaci species identified in this study. There was a clear difference in the distribution of whiteflies in the different agro-ecological zones. SSA1-SG1 was the most widely distributed B. tabaci species in the nine agro-ecologies where whiteflies were collected (Figure 4A); followed by MED-ASL that occurred in seven agro-ecologies (Figure 4D). SSA1-SG1 dominated in the north-eastern drylands, Kyoga plains, western savanna grassland, south-western farmland and highland ranges comprising of 50% (7/14), 45.6% (31/68), 42.6% (20/47), 34.8% (8/23) and 37.8% (17/45) of the total whiteflies collected in each agro-ecological zone, respectively, whereas MED-ASL predominated in north-western savanna grassland and Lake Victoria crescent with 76.9% (10/13) and 33.3% (27/81), respectively (Table 2). IO was the most predominant in the north-eastern savanna grassland consisting of 34.3% (12/35) (Table 2). SSA1-SG1 and MED-Q1 occurred equally in the para savanna, (1/2) each. MED-Q1 was found in six agro-ecologies (Table 2; Figure 4C). The SSA1-SG2 and SSA6 were distributed in five agro-ecologies (Table 2; Figures 4A, E). SSA11 and SSA13 were found in four agro-ecologies (Table 2; Figures 4C, E). Three of the B. tabaci species namely SSA1-Hoslundia, SSA2 and IO were distributed in three agro-ecologies (Table 2; Figures 4B, E). Only SSA12 was distributed in two agro-ecologies (Table 2; Figure 4C). Four of the B. tabaci species; SSA1-SG3, SSA3, SSA10 and MEAM1 occurred in only one agro-ecological region (Table 2; Figures 4B–D). Other whiteflies identified in this study were B. afer and B. Uganda1 and were distributed in six agro-ecologies (Table 2; Figure 4F).
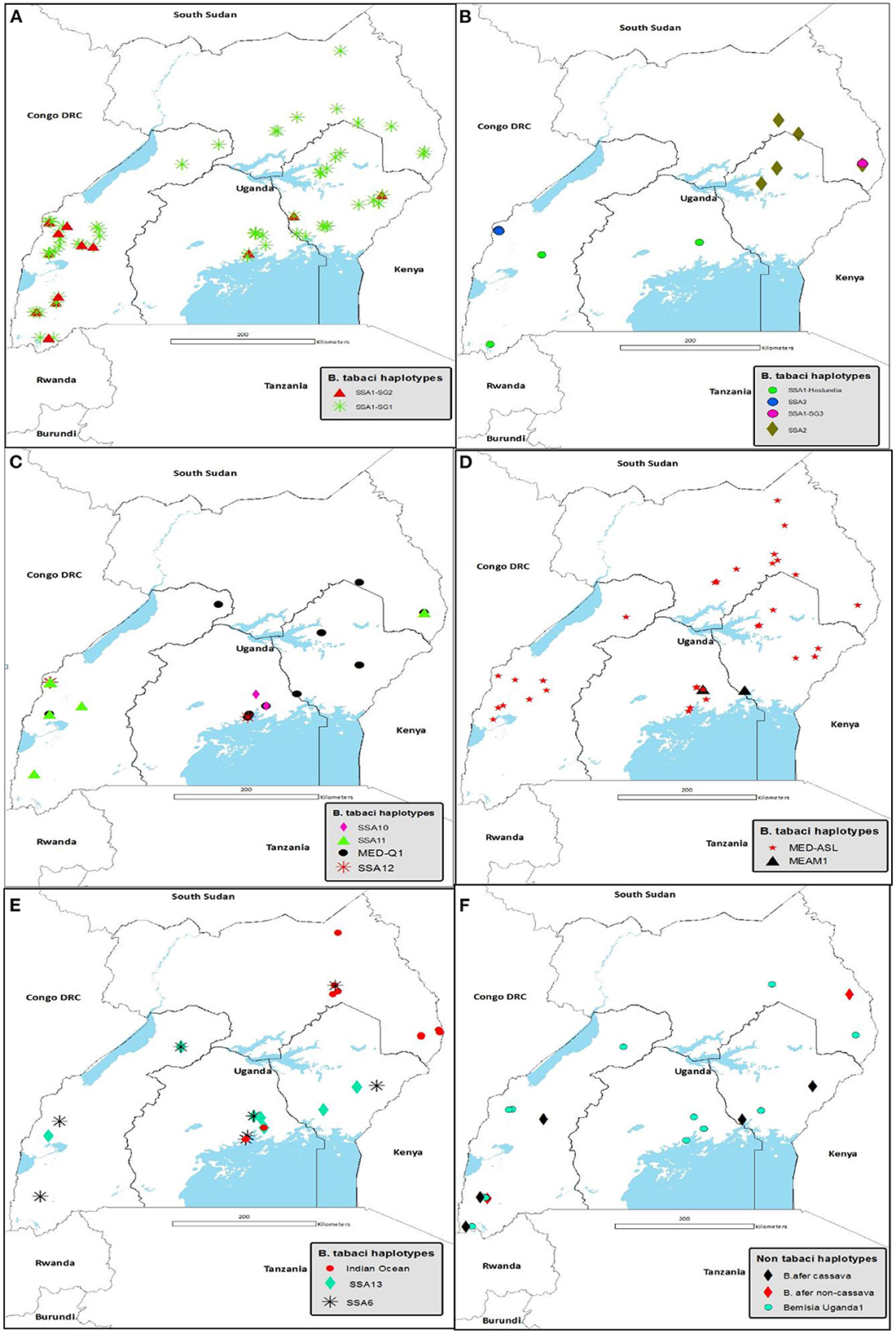
Figure 4. Agro-ecological distribution of (A) SSA1-SG1 and SSA1-SG2: (B) SSA1-Hoslundia, SSA1-SG3, SSA2 and SSA3: (C) SSA10, SSA11, SSA12 and MED-Q1: (D) MED-ASL and MEAM1: (E) SSA6, SSA13 and Indian Ocean: and (F) B. afer and Bemisia Uganda1 species collected in Uganda during May/August 2017. Abbreviations for the B. tabaci species: SSA, sub-Saharan Africa; SG, sub-group; MED, Mediterranean; ASL, African silver leafing and MEAM1, Middle East-Asia Minor1.
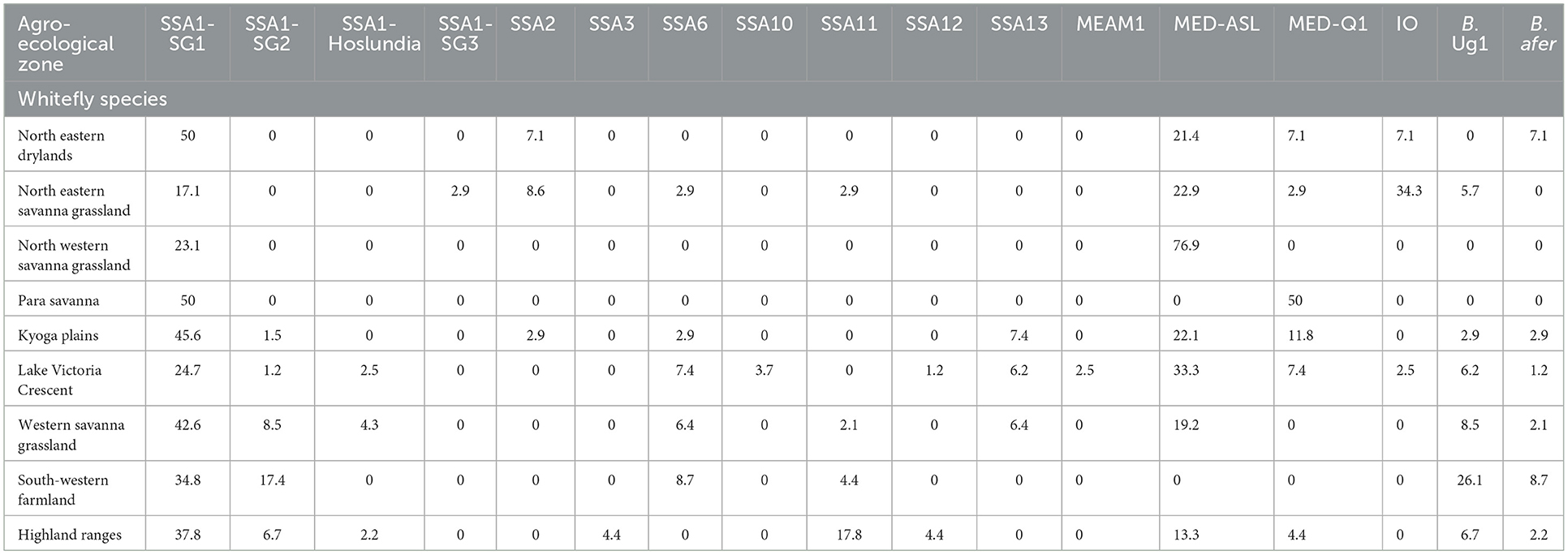
Table 2. Percentage composition of adult whiteflies collected in the different agro-ecologies during May/August 2017 in Uganda.
Discussion
Cassava is a key food security crop in many sub-Saharan African countries, but production is affected by pests and viral diseases. B. tabaci is a pest and a vector of cassava viruses, so it is important to understand B. tabaci species and their distributions in the different agro-ecological zones in order to deploy management strategies appropriately. In this study, we conducted an extensive survey in the different agro-ecological zones that included cassava growing areas and areas with limited or no cassava growing. This study examined 238 good-quality sequences for whitefly individuals collected in nine agro-ecological zones from 34 host-plants in Uganda.
Bemisia tabaci species identified on cassava
The B. tabaci species identified on cassava were: SSA1 (-SG1, -SG2 and -Hoslundia) (88.3%), SSA2 (2.7%), SSA3 (0.9%), MED-ASL (2.7%) and IO (0.9%). SSA1-SG1 (75.7%) dominated all the whiteflies identified on cassava. Of the five B. tabaci species, SSA1, SSA2, SSA3 have been reported to colonize cassava in sub-Saharan Africa (Sseruwagi et al., 2005; Legg et al., 2014; Ghosh et al., 2015; Nwezeobi et al., 2020; Mugerwa et al., 2021b). SSA1-SG1, SSA1-SG2 and SSA2 were reported as the main cassava colonizers in Uganda and Kenya (Mugerwa et al., 2012, 2021a; Legg et al., 2014), while SSA1-SG1 and SSA1-SG3 were the major colonizers of cassava in Tanzania (Ghosh et al., 2015; Tajebe et al., 2015). Results of our study complement earlier studies that reported the presence of SSA1 (-SG1, -SG2 and -Hoslundia) and SSA2 on cassava in Uganda (Legg et al., 2014). Nonetheless, SSA1-SG3 was found on a non-cassava host-plant, P. vulgaris. SSA1-SG3 was not detected on cassava, probably because it was rare or it has been displaced to alternative host-plants by the high populations of SSA1-SG1. Similarly, Mugerwa et al. (2021a) found SSA1-SG3 on alternative host-plants and these included; P. vulgaris, S. melongena, H. opposita, Ageratum sp. and S. acuta. In addition, Mugerwa et al. (2021b) observed that SSA1-SG3 was the least fecund amongst the three sub-groups (-SG1, -SG2 and -SG3) of SSA1 species. Recent findings by Mugerwa et al. (2020) showed that there was mating compatibility between SSA1-SG1 and SSA1-SG2, while SSA1-SG3 and SSA1-SG1 were reproductively incompatible, which possibly explains why SSA1-SG3, a different species from SSA1-SG1 and SSA1-SG2, differs in its occurrence on cassava and fecundity. It is thus likely that the high populations of SSA1-SG1 have displaced SSA1-SG3 from cassava to alternative hosts in Uganda.
The SSA1-SG1 and SSA2 are reported as associated with the spread of cassava viruses that cause CMD and CBSD, which cause up to 50% yield losses in East Africa (Legg et al., 2002, 2014; Maruthi et al., 2005). From 2000, it was reported that SSA2 was gradually replaced by SSA1-SG1 (Legg et al., 2014), and the high populations of whiteflies on cassava and spread of CMD were attributed to SSA1-SG1 (Mugerwa et al., 2012; Legg et al., 2014; Tajebe et al., 2015), which was also the most predominant SSA1 species in this study.
We report here for the first time the presence of SSA3 in Uganda. SSA3 is one of the key B. tabaci species colonizing cassava in Nigeria (Ghosh et al., 2015; Nwezeobi et al., 2020). Nwezeobi et al. (2020) reported that SSA3 has been found in eastern Nigeria, Benin, Cameroon, Togo and Central Africa Republic. Legg et al. (2014) found SSA3 in the western part of DRC unaffected by the CMD pandemic. This area shares boundaries with the district where our study identified SSA3. Therefore, SSA3 could have probably been introduced to East Africa from West Africa. Although SSA3 was widely distributed in eastern Nigeria, it was in low numbers and did not pose a significant threat to cassava in terms of direct damage (Nwezeobi et al., 2020). In addition, CMD incidence in the field was not as severe and widely distributed as the current CMD epidemic in Eastern Africa (Nwezeobi et al., 2020). This suggests that SSA3 may not be a serious threat as pest and vector as SSA1-SG1. The MED-ASL and IO species occurred on cassava, but earlier studies that involved collecting fourth instar nymphs reported that none of these colonized cassava (Sseruwagi et al., 2005). The MED-ASL and IO, therefore, were probably transient adult whiteflies on the cassava.
Cassava B. tabaci occurred on other host-plants
The cassava B. tabaci species identified were SSA1, SSA2 and SSA3. Of these, SSA1 particularly SSA1-SG1 was the most polyphagous, occurring on 18 plant species. Previous studies reported that SSA1-SG1 occurred on more than 30 host-plant species compared to other subgroups, -SG2 and -SG3 (Tocko-Marabena et al., 2017; Mugerwa et al., 2021a). This suggests that SSA1-SG1 utilizes these host-plants in the absence of cassava, hence favoring its population growth. Although SSA1-SG1 was polyphagous, it was commonly found on cassava, which was in agreement with the findings by Mugerwa et al. (2021a). Otim et al. (2018) did not differentiate between whitefly species but also reported that B. tabaci and its parasitoids were more abundant on cassava than intercrops and weeds, an indication that cassava was the preferred host for cassava B. tabaci.
The ability of cassava B. tabaci to feed on various host-plant species increases the risk of transmitting viruses between these host-plants. In a review by García-Arenal and Zerbini (2019), they proposed that spread of viral diseases from their original wild host-plants to crops is done by polyphagous whitefly vector populations. Ramkat et al. (2011) identified two cassava mosaic geminiviruses, ACMV and EACMV-UG in J. carcus in Kenya and Ethiopia. Alabi et al. (2008) reported the presence of ACMV from Senna occidentalis, Leucana leucocephala, Glycine max, Ricinus communis, Combretum confertum and M. glaziovii found in close proximity with cassava in Nigeria. Transmission of viruses between plants may lead to evolution of new viruses through recombination (Seal et al., 2006), which can result in new pandemics that are more devastating; recombination between two cassava viruses (ACMV and EACMV) resulted in a more severe virus strain, EACMV-UG that caused a more devastating CMD pandemic in Uganda in the 1990s (Zhou et al., 1997; Pita et al., 2001).
Distribution of cassava B. tabaci in different agro-ecological zones
This study revealed the importance of sampling a broad geographic area to include almost all agro-ecological zones. The Karamoja sub-region had never been sampled due to fear of hostility. The Karamojong are mainly pastoralists, who practice cattle rustling because cattle are considered as wealth (Odhiambo, 2003; Bevan, 2008). Similarly, no sampling had been done in the highland ranges and southwestern farmland due to the steep terrain and being dominated by banana growing, respectively. The SSA1-SG1 was widely distributed in the nine agro-ecologies. SSA1-SG1 is polyphagous (Sseruwagi et al., 2006; Mugerwa et al., 2021a), which possibly explains its ability to exist in all agro-ecological zones. It was detected on J. curcas in Kaabong, where there was no cassava and on E. heterophylla and the few cassava plants found in Napak, both districts are found in the north-eastern drylands in the Karamoja sub-region. The existence of SSA1-SG1 in diverse agro-ecological regions on cultivated and non-cultivated plants will enable this species to easily be reintroduced to targeted control areas from neighboring fields by wind or on plant material by humans. Effective management should therefore focus on resistance to cassava whiteflies to reduce the high vector populations and associated viral disease outbreaks. There are considerable research efforts targeting resistance to cassava whiteflies (http://www.cassavawhitefly.org) and it is hoped these efforts will be transferable in the near future to facilitate breeding for resistance to prevalent whitefly species impacting other crops in sub-Saharan Africa. Kaabong district is within the semi-arid zone mainly composed of pastoral activities and crops grown include millet, cowpeas and groundnuts, whereas Napak district is an agro-pastoral area, growing sorghum, maize, beans, sesame, sunflower, vegetables and fruits (Kraybill and Kidoido, 2009; USAID, 2017b; FAO, 2018). In an effort to increase food production in these areas, the government of Uganda in collaboration with Food and Agriculture Organization of United Nations (FAO) introduced irrigation and seed multiplication sites for sweetpotato and cassava (FAO, 2013). SSA1-SG1 was also found in the south-western farmlands in Bushenyi and Rukungiri districts and highland ranges in Bulambuli and Kabale districts, where the main crops grown are bananas and potatoes, respectively. The presence of SSA1-SG1 in areas with no or limited cassava growing could be explained by presence of host-plants where it existed before the introduction of cassava into East Africa in the eighteenth century (Guthrie, 1987). Recent studies have shown that Africa is the center of origin of B. tabaci based on the estimated dates of divergence (Mugerwa et al., 2018). It is likely that cassava B. tabaci has evolved the ability to colonize cassava, which probably led to development of populations with preference for cassava.
Contrary to SSA1-SG1, the SSA2 species was found in the north-eastern drylands and north-eastern savanna grassland in Karamoja sub-region and the Kyoga plains in eastern Uganda. The three agro-ecologies are characterized by short savanna grassland (Mugerwa et al., 2012), with Karamoja having unimodal rainfall while the Kyoga plains have bimodal rainfall (Kraybill and Kidoido, 2009). Mugerwa et al. (2021a) reported that SSA2 was more prevalent in northern Uganda, which shares a similar agro-ecology with the areas where this study identified it. Northern Uganda neighbors South Sudan, where Misaka et al. (2020) found that SSA2 was the most prevalent. The difference in distribution between SSA1-SG1 and SSA2 also partly explains the change in the B. tabaci species associated with CMD over time. It was reported that SSA2 was responsible for the spread of the CMD pandemic from northern and eastern Uganda to southern Uganda in the 1990s (Legg et al., 2002). However, from 2000 to present CMD has been associated with SSA1-SG1 (Sseruwagi et al., 2006; Mugerwa et al., 2012; Legg et al., 2014) that has a wider agro-ecological distribution. In addition, SSA2 was found to be more predominant than SSA1-SG1 in western Kenya a CMD pandemic area in 2001 (Legg et al., 2014), which neighbors eastern Uganda. Recently, Macfadyen et al. (2020) found SSA2 in northern and eastern Uganda, an indication that it is affected by agro-ecology and landscape factors. SSA3 was also found in Bundibugyo district, neighboring eastern Democratic Republic of Congo (DRC), where it was previously reported to occur (Legg et al., 2014) and they share the same agro-ecology.
The SSA1-SG2, although having been confirmed to be reproductively compatible with SSA1-SG1 (Mugerwa et al., 2020), was not as widely distributed as SSA1-SG1. However, it was also found in cassava growing areas in the Lake Victoria crescent, Kyoga plains and western savanna grassland and non-cassava growing areas in the highland ranges and south-western farmlands. This complements earlier findings that found SSA1-SG2 in cassava growing areas in the Lake Victoria crescent, and non-cassava growing areas in the pastoral rangeland in Uganda (Mugerwa et al., 2021a), the southern part of Ukerewe Island in Tanzania, the northern part of Western Kenya (Legg et al., 2014), the coast province in Kenya and the coast and southern zones in Tanzania (Mugerwa et al., 2012). In this study and earlier studies by Mugerwa et al. (2012) and Mugerwa et al. (2021a), SSA1-SG2 was not detected in areas characterized by unimodal type of rainfall in Uganda, suggesting better adaptation to areas with bimodal rainfall.
Implications of cassava B. tabaci existence on alternative hosts in new cassava growing areas for the control of whitefly-transmitted viruses
Cassava begomoviruses are endemic to Africa (Tiendrébéogo et al., 2012) and wild host-plants acts as reservoirs of these viruses (Alabi et al., 2008; Ramkat et al., 2011; Amisse et al., 2019). Therefore occurrence of SSA1-SG1 on cultivated and non-cultivated plants could increase the acquisition and transmission of begomoviruses between these plants. In addition, if susceptible cassava varieties are introduced into non-cassava growing areas like Kaabong where SSA1-SG1 was found on J. curcas that is a wild host plant of cassava mosaic geminiviruses (Ramkat et al., 2011), they may possibly be infected by viruses from alternative hosts, which could lead to virus outbreaks. Rey et al. (2012) also reviewed and concluded that on the African continent and proximal Indian Ocean islands, outbreaks of begomovirus diseases are possibly due to the introduction and intensive cultivation of exotic crops in environments with indigenous begomoviruses. These viruses are transmitted by polyphagous B. tabaci between natural hosts and introduced plant species.
Non-cassava B. tabaci identified in the study
The non-cassava B. tabaci identified in the study were: MED (-ASL and -Q1), MEAM1, IO, SSA6, SSA10, SSA11, SSA12 and SSA13. There was also a clear distinction in host-plant range and agro-ecological distribution amongst the different whitefly species. MED-ASL was the most predominant on non-cassava host-plants. Although MED-Q1 has been considered the same species as MED-ASL based on the partial mtCO1 (De Barro et al., 2011), Vyskočilová et al. (2018) demonstrated that Uganda MED-ASL and MED-Q1 are reproductively incompatible and thus different biological species. In this study, differences between MED-Q1 and MED-ASL were observed in their occurrence on N. tabacum, and with MED-ASL being more widely distributed across different agro-ecologies than MED-Q1. These results are in agreement with findings by Mugerwa et al. (2021a) who reported that MED-ASL was the most predominant of the non-cassava B. tabaci in Uganda, and that MED-Q1 was found more frequently on N. tabacum than MED-ASL.
MEAM1 and MED species are known to be global invasive species (De Barro et al., 2011; Hu et al., 2011). There have been reports of MEAM1 displacing indigenous B. tabaci species in different parts of the world (Costa and Brown, 1991; Liu et al., 2007) and MED displacing MEAM1 (Chu et al., 2010). In this study, MEAM1 was found on only two plant species in the Lake Victoria crescent, which is in agreement with earlier findings that MED was more prevalent than MEAM1 in Uganda (Sseruwagi et al., 2005; Mugerwa et al., 2021a). The SSA6 was found only on O. gratissimum, which concurred with results of Sseruwagi et al. (2005). Contrary to findings by Mugerwa et al. (2018) who reported presence of SSA10 on cassava, this whitefly species occurred only on V. amygdalina in this study.
Conclusions
There is a high diversity of B. tabaci species in Uganda, but a clear distinction in host-plant range and agro-ecological distribution amongst the different B. tabaci species found [SSA1 (-SG1, -SG2, -SG3 and -Hoslundia), SSA2, SSA3, SSA6, SSA10, SSA11, SSA12, SSA13, IO, MEAM1 and MED (-ASL and -Q1)]. In addition, there is strong evidence that some B. tabaci species, such as SSA1-SG2, SSA2 and SSA3, are restricted in their distributions due to the agro-ecologies present in Uganda. SSA1-SG1 and MED-ASL were found on many cultivated and non-cultivated plant species. SSA1-SG1 was the most polyphagous cassava B. tabaci and was widely distributed in the different agro-ecological zones. It was found in major cassava growing areas and areas where little or no cassava was grown. Information about the wide distribution of SSA1-SG1 is important when distributing planting materials and deploying pest management strategies. Measures must be put in place to prevent introduction of diseased materials into new areas, since the vectors that can spread the viruses exist. Also, farmers should be provided with disease resistant materials to minimize the possibility of transmission of viruses from alternative hosts.
Furthermore, the continuous high whitefly populations observed on cassava in Uganda could be due to the ability of SSA1-SG1 to adapt to different climatic conditions and cropping systems and also utilize other host-plants when cassava is harvested, which probably favors its population growth. Also, MED-ASL, an important invasive species of B. tabaci that transmits viruses to ornamental, horticultural crops and other agricultural crops was also widely spread and highly polyphagous in Uganda. Therefore, this information is required when developing and deploying appropriate pest and disease management strategies internationally. If MED-ASL were to spread out of its home range in sub-Saharan Africa, for example, it is clearly polyphagous enough to become established in new geographical regions.
Data availability statement
All the 66 unique haplotype sequences of the whiteflies identified in this study are available in the GenBank sequence database (accession numbers MN271483 – MN271548).
Author contributions
SS, CO, SM, and JC conceptualized the work and acquired funding. AN and RK took part in data collection. AN, SB, and JC participated in data analysis. CO, OM, SM, SS, and JC were involved in supervision. AN drafted the original manuscript. All authors contributed to the article.
Funding
This work was funded by the Bill and Melinda Gates Foundation through the African Cassava Whitefly Project [Grant Agreement OPP1058938]. Under the grant conditions of the Foundation, a Creative Commons Attribution 4.0 Generic License has already been assigned to the Author Accepted Manuscript version that might arise from this submission.
Acknowledgments
We thank the farmers who gave us access to collect whitefly from their fields. We are also grateful to Dr. Meredith Williams on his advice on generating maps using the ArcMap software and acknowledge Mr. Phillip Abidrabo for drawing the map of Uganda with different agro-ecological regions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alabi, O. J., Kumar, P. L., and Naidu, R. A. (2011). Cassava mosaic disease: A curse to food security in sub-Saharan Africa. APSnet Features. Washington: APS, 1–16.
Alabi, O. J., Ogbe, F. O., Bandyopadhyay, R., Kumar, P. L., Dixon, A. G. O., Hughes, J. d'A., et al. (2008). Alternate hosts of African cassava mosaic virus and East African cassava mosaic Cameroon virus in Nigeria. Arch. Virol. 153, 1743–1747. doi: 10.1007/s00705-008-0169-8
Alicai, T., Ndunguru, J., Sseruwagi, P., Tairo, F., Okao-Okuja, G., Nanvubya, R., et al. (2016). Cassava Brown Streak Virus Has a Rapidly Evolving Genome: Implications for Virus Speciation, Variability, Diagnosis and Host Resistance. Sci. Rep. 6, 36164. doi: 10.1038/srep36164
Alicai, T., Omongo, C. A., Maruthi, M. N., Hillocks, R. J., Baguma, Y., Kawuki, R., et al. (2007). Re-emergence of cassava brown streak disease in Uganda. Plant Dis. 91, 24–29. doi: 10.1094/PD-91-0024
Alicai, T., Szyniszewska, A. M., Omongo, C. A., Abidrabo, P., Okao-Okuja, G., Baguma, Y., et al. (2019). Expansion of the cassava brown streak pandemic in Uganda revealed by annual field survey data for 2004 to 2017. Sci Data 6, 327. doi: 10.1038/s41597-019-0334-9
Ally, H. M., El Hamss, H., Simiand, C., Maruthi, M. N., Colvin, J., Omongo, C. A., et al. (2019). What has changed in the outbreaking populations of the severe crop pest whitefly species in cassava in two decades? Sci. Rep. 9, 14796. doi: 10.1038/s41598-019-50259-0
Amisse, J. J. G., Ndunguru, J., Tairo, F., Boykin, L. M., Kehoe, M. A., Cossa, N., et al. (2019). First report of cassava brown streak viruses on wild plant species in Mozambique. Physiol. Mol. Plant Pathol. 105, 88–95. doi: 10.1016/j.pmpp.2018.10.005
Ayres, D. L., Darling, A., Zwickl, D. J., Beerli, P., Holder, M. T., Lewis, P. O., et al. (2012). BEAGLE: An application programming interface and high-performance computing library for statistical phylogenetics. Syst. Biol. 61, 70–173. doi: 10.1093/sysbio/syr100
Berry, S. D., Fondong, V. N., Rey, C., Rogan, D., Fauquet, C. M., Brown, J. K., et al. (2004). Molecular evidence for five distinct Bemisia tabaci (Homoptera:Aleyrodidae) geographic haplotypes associated with cassava plants in sub-Saharan Africa. BioOne 3, 852–859. doi: 10.1603/0013-8746(2004)097[0852:MEFFDB]2.0.CO;2
Bevan, J. (2008). Crisis in Karamoja: Armed violence and the failure of disarmament in Uganda's most deprived region. Small Arms Survey, Occassional Paper 21, 20–23.Available at: https://www.jstor.org/stable/resrep10750.12
Brown, J. K., Frohlich, D. R., and Rosell, R. C. (1995). The sweetpotato or silverleaf whiteflies: Biotypes of Bemisia tabaci or a species complex? Annu. Rev. Entomol. 40, 511–534. doi: 10.1146/annurev.en.40.010195.002455
Brown, J. K., Zerbini, F. M., Navas-Castillo, J., Moriones, E., Ramos-Sobrinho, R., Silva, J. C., et al. (2015). Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch. Virol. 160, 1593–1619. doi: 10.1007/s00705-015-2398-y
Bull, S. E., Briddon, R. W., Sserubombwe, W. S., Ngugi, K. G., Markham, P. G., and Stanley, J. (2006). Genetic diversity and phylogeography of cassava mosaic viruses in Kenya. J. Gen. Virol. 87, 3053–3065. doi: 10.1099/vir.0.82013-0
Burns, A., Gleadow, R., Cliff, J., Zacarias, A., and Cavagnaro, T. (2010). Cassava: The drought, war and famine crop in a changing world. Sustainability 2, 3572–3607. doi: 10.3390/su2113572
Butler, G. D., and Henneberry, T. J. (1985). Bemisia tabaci (Gennadius), a pest of cotton in the Southwestern United States. U.S. Depart. Agricult. Tech. Bull. 1707, 1–19. doi: 10.22004/ag.econ.156822
Byrne, D. N., and Bellows, T. S. (1991). Whitefly biology. Annu. Rev. Entomol. 36, 431–457. doi: 10.1146/annurev.en.36.010191.002243
Chu, D., Wan, F. H., Zhang, Y. J., and Brown, J. K. (2010). Change in the biotype composition of Bemisia tabaci in Shandong Province of China from 2005 to 2008. Environ. Entomol. 39, 1028–1036. doi: 10.1603/EN09161
Colvin, J., Omongo, C. A., Maruthi, M. N., Otim-nape, G. W., and Thresh, J. M. (2004). Dual begomovirus infections and high Bemisia tabaci populations: two factors driving the spread of a cassava mosaic disease pandemic. Plant Pathol. 1, 577–584. doi: 10.1111/j.0032-0862.2004.01062.x
Costa, H. S., and Brown, J. K. (1991). Variation in biological characteristics and esterase patterns among populations of Bemisia tabaci, and the association of one population with silverleaf symptom induction. Entomol. Exp. Appl. 61, 211–219. doi: 10.1111/j.1570-7458.1991.tb01553.x
De Barro, P. J., Liu, S., Boykin, L. M., and Dinsdale, A. B. (2011). Bemisia tabaci : A statement of species status. Annu. Rev. Entomol. 56, 1–19. doi: 10.1146/annurev-ento-112408-085504
Dinsdale, A., Cook, L., Riginos, C., Buckley, Y. M., and De Barro, P. (2010). Refined global analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) mitochondrial cytochrome oxidase 1 to identify species level genetic boundaries. Ann. Entomol. Soc. Am. 103, 196–208. doi: 10.1603/AN09061
El-sharkawy, M. A. (2007). Physiological characteristics of cassava tolerance to prolonged drought in the tropics : Implications for breeding cultivars adapted to seasonally dry and semiarid environments. Braz. J. Plant Physiol. 19, 257–286. doi: 10.1590/S1677-04202007000400003
Esterhuizen, L. L., Mabasa, K. G., Van Heerden, S. W., Czosnek, H., Brown, J. K., Van Heerden, H., et al. (2013). Genetic identification of members of the Bemisia tabaci cryptic species complex from South Africa reveals native and introduced haplotypes. J. Appl. Entomol. 137, 122–135. doi: 10.1111/j.1439-0418.2012.01720.x
FAO (2013). Save and Grow: Cassava. A Guide to Sustainable Production Intensification. Québec City, Canada: Food and Agriculture Organization of the United Nations.
FAO (2018). Analysing Resilience for Better Targeting and Action: Resilience Analysis in Karamoja, Uganda. Québec City, Canada: Food and Agricultural Organisation of United Nations Resilience Analysis Report No. 10.
FAO (2019). Protecting Cassava, a Neglected Crop from Pests and Diseases. Québec City, Canada: Food and Agriculture Organization of the United Nations. www.fao.org/food-chain-crisis
García-Arenal, F., and Zerbini, F. M. (2019). Life on the Edge: Geminiviruses at the interface between crops and wild plant hosts. Annu. Rev. Virol. 6, 411–433. doi: 10.1146/annurev-virology-092818-015536
Ghosh, S., Bouvaine, S., and Maruthi, M. N. (2015). Prevalence and genetic diversity of endosymbiotic bacteria infecting cassava whiteflies in Africa. BMC Microbiol. 15, 1–17. doi: 10.1186/s12866-015-0425-5
Gilbertson, R. L., Batuman, O., Webster, C. G., and Adkins, S. (2015). Role of the insect supervectors Bemisia tabaci and Frankliniella occidentalis in the emergence and global spread of plant viruses. Annu. Rev. Virol. 2, 67–93. doi: 10.1146/annurev-virology-031413-085410
Guthrie, E. J. (1987). “African cassava mosaic disease and its control,” in International Seminar on African Cassava Mosaic Disease and its Control. Yamoussoukro, Ivory coast, 1–9.
Hu, J., De Barro, P., Zhao, H., Wang, J., Nardi, F., and Liu, S. S. (2011). An extensive field survey combined with a phylogenetic analysis reveals rapid and widespread invasion of two alien whiteflies in China. PLoS ONE 6, e16061. doi: 10.1371/journal.pone.0016061
Jarvis, A., Ramirez-Villegas, J., Campo, B. V. H., and Navarro-Racines, C. (2012). Is cassava the answer to African climate change adaptation? Trop. Plant Biol. 5, 9–29. doi: 10.1007/s12042-012-9096-7
Jones, D. R. (2003). Plant viruses transmitted by white?ies. Eur. J. Plant Pathol. 109, 195–219. doi: 10.1023/A:1022846630513
Joyce, R. J. V. (1983). Aerial transport of pests and pest outbreaks. EPPO Bull. 13, 111–119. doi: 10.1111/j.1365-2338.1983.tb01585.x
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. E30, 772–780. doi: 10.1093/molbev/mst010
Kawuki, R. S., Kaweesi, T., Esuma, W., Pariyo, A., Kayondo, I. S., Ozimati, A., et al. (2016). Eleven years of breeding efforts to combat cassava brown streak disease. Breed. Sci. 66, 560–571. doi: 10.1270/jsbbs.16005
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kraybill, D., and Kidoido, M. (2009). Analysis of relative profitability of key Ugandan agricultural enterprises by agricultural production zone. Uganda: International Food Policy Research Institute, Uganda Strategy Support Program (USSP), Background Paper no. USSP 04 pp. 2–5.
Kunz, D., Tay, W. T., Elfekih, S., Gordon, K. H. J., and De Barro, P. J. (2019). Take out the rubbish–Removing NUMTs and pseudogenes from the Bemisia tabaci cryptic species mtCOI database. bioRxiv. doi: 10.1101/724765
Lecoq, H., Verdin, E., Tepfer, M., Wipf-Scheibel, C., Millot, P., Dafalla, G., et al. (2016). Characterization and occurrence of squash chlorotic leaf spot virus, a tentative new torradovirus infecting cucurbits in Sudan. Arch. Virol. 161, 1651–1655. doi: 10.1007/s00705-016-2797-8
Legg, J. P. (1994). Bemisia tabaci: the whitefly vector of cassava mosaic geminiviruses in Africa: An ecological perspective. Afr. Crop Sci. J. 2, 437–448.
Legg, J. P., French, R., Rogan, D., Okao-Okuja, G., and Brown, J. K. (2002). A distinct Bemisia tabaci (Gennadius) (Hemiptera: Sternorrhyncha: Aleyrodidae) genotype cluster is associated with the epidemic of severe cassava mosaic virus disease in Uganda. Mol. Ecol. 11, 1219–1229. doi: 10.1046/j.1365-294X.2002.01514.x
Legg, J. P., Jeremiah, S. C., Obiero, H. M., Maruthi, M. N., Ndyetabula, I., Okao-Okuja, G., et al. (2011). Comparing the regional epidemiology of the cassava mosaic and cassava brown streak virus pandemics in Africa. Virus Res. 159, 161–170. doi: 10.1016/j.virusres.2011.04.018
Legg, J. P., Sseruwagi, P., Boniface, S., Okao-Okuja, G., Shirima, R., Bigirimana, S., et al. (2014). Spatio-temporal patterns of genetic change amongst populations of cassava Bemisia tabaci whiteflies driving virus pandemics in East and Central Africa. Virus Res. 186, 61–75. doi: 10.1016/j.virusres.2013.11.018
Liu, S. S., Colvin, J., and De Barro, P. J. (2012). Species concepts as applied tp the whitefly Bemisia tabaci systematics: How many species are there? J. Integr. Agric. 11, 176–186. doi: 10.1016/S2095-3119(12)60002-1
Liu, S. S., De Barro, P. J., Xu, J., Luan, J. B., Zang, L. S., Ruan, Y. M., et al. (2007). Asymmetric mating interactions drive widespread invasion and displacement in a whitefly. Science 318, 1769–1772. doi: 10.1126/science.1149887
Macfadyen, S., Tay, W. T., Hulthen, A. D., Paull, C., Kalyebi, A., Jacomb, F., et al. (2020). Landscape factors and how they influence whitefly pests in cassava fields across East Africa. Landsc. Ecol. 36, 45–67. doi: 10.1007/s10980-020-01099-1
Manani, D. M., Ateka, E. M., Nyanjom, S. R. G., and Boykin, L. M. (2017). Phylogenetic relationships among whiteflies in the Bemisia tabaci (Gennadius) species complex from major cassava growing areas in Kenya. Insects 8, 1–14. doi: 10.3390/insects8010025
Maruthi, M. N., Colvin, J., Seal, S., Gibson, G., and Cooper, J. (2002). Co-adaptation between cassava mosaic geminiviruses and their local vector populations. Virus Res. 86, 71–85. doi: 10.1016/S0168-1702(02)00051-5
Maruthi, M. N., Hillocks, R. J., Mtunda, K., Raya, M. D., Muhanna, M., Kiozia, H., et al. (2005). Transmission of cassava brown streak virus by Bemisia tabaci (Gennadius). J. Phytopathol. 153, 307–312. doi: 10.1111/j.1439-0434.2005.00974.x
Mbanzibwa, D. R., Tian, Y. P., Tugume, A. K., Mukasa, S. B., Tairo, F., Kyamanywa, S., et al. (2009). Genetically distinct strains of cassava brown streak virus in the Lake Victoria basin and the Indian Ocean coastal area of East Africa. Arch. Virol. 154, 353–359. doi: 10.1007/s00705-008-0301-9
Misaka, B. C., Wosula, E. N., Marchelo-d'Ragga, P. W., Hvoslef-Eide, T., and Legg, P. J. (2020). Genetic diversity of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) colonizing sweetpotato and cassava in South Sudan. Insects 11, 58. doi: 10.3390/insects11010058
Monger, W. A., Seal, S., Isaac, A. M., and Foster, G. D. (2001). Molecular characterization of the cassava brown streak virus coat protein. Plant Pathol. 50, 527–534. doi: 10.1046/j.1365-3059.2001.00589.x
Mugerwa, H., Colvin, J., Alicai, T., Omongo, C. A., Kabaalu, R., Visendi, P., et al. (2021a). Genetic diversity of whitefly (Bemisia spp.) on crop and uncultivated plants in Uganda: implications for the control of this devastating pest species complex in Africa. J. Pest Sci. 1–24, doi: 10.1007/s10340-021-01355-6
Mugerwa, H., Rey, M. E. C., Alicai, T., Ateka, E., Atuncha, H., Ndunguru, J., et al. (2012). Genetic diversity and geographic distribution of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) genotypes associated with cassava in East Africa. Ecol. E2, 2749–2762. doi: 10.1002/ece3.379
Mugerwa, H., Seal, S., Wang, H.-. L., Patel, M. V., Kabaalu, R., Omongo, C. A., et al. (2018). African ancestry of New World, Bemisia tabaci-whitefly species. Sci. Rep. 8, 2734. doi: 10.1038/s41598-018-20956-3
Mugerwa, H., Sseruwagi, P., Colvin, J., and Seal, S. (2021b). Is high whitefly abundance on cassava in sub-Saharan Africa driven by biological traits of a specific, cryptic Bemisia tabaci species? Insects, 12, 260. insects12030260. doi: 10.3390/insects12030260
Mugerwa, H., Wang, H. L., Sseruwagi, P., Seal, S., and Colvin, J. (2020). Whole-genome single nucleotide polymorphism and mating compatibility studies reveal the presence of distinct species in sub-Saharan Africa Bemisia tabaci whiteflies. Insect Sci. 28, 1553–1566. doi: 10.1111/1744-7917.12881
Namuddu, A. (2019). Ecological and genetic factors contributing to high cassava whitefly, Bemisia tabaci, populations in East Africa (PhD thesis). University of Greenwich (UK).
Ndunguru, J., Sseruwagi, P., Tairo, F., Stomeo, F., Maina, S., Djikeng, A., et al. (2015). Analyses of Twelve New Whole Genome Sequences of Cassava Brown Streak Viruses and Ugandan Cassava Brown Streak Viruses from East Africa: Diversity, Supercomputing and Evidence for Further Speciation. PLoS ONE 10, e0139321. doi: 10.1371/journal.pone.0139321
Nwezeobi, J., Onyegbule, O., Nkere, C., Onyeka, J., van Brunschot, S., Seal, S., et al. (2020). Cassava whitefly species in eastern Nigeria and the threat of vector-borne pandemics from East and Central Africa. PLoS ONE 15, e0232616. doi: 10.1371/journal.pone.0232616
Odhiambo, M. O. (2003). The Karamoja Conflict: Origins, Impact and Solutions. @ Oxfam, Fountain Publishers Ltd, 1–75.
Oliveira, M. R. V., Henneberry, T. J., and Anderson, P. (2001). History, current status, and collaborative research projects for Bemisia tabaci. Crop Prot. 20, 709–723. doi: 10.1016/S0261-2194(01)00108-9
Omongo, C. A., Kawuki, R., Bellotti, A. C., Alicai, T., Baguma, Y., Maruthi, M. N., et al. (2012). African cassava whitefly, Bemisia tabaci, resistance in African and South American cassava genotypes. J. Integr. Agric. 11, 327–336. doi: 10.1016/S2095-3119(12)60017-3
Otim, M., Kyamanywa, S., Ecaat, S., Legg, J., and Gerling, D. (2018). The incidence of Bemisia tabaci (Homoptera: Aleyrodidae) and its parasitoids on cassava and associated plants in Uganda. Isr. J. Entomol. 48, 157–176.
Otim-Nape, G. W., Bua, A., and Baguma, Y. (1994). Accelerating the transfer of improved production technologies: Controlling African cassava mosaic virus disease epidemics in Uganda. Afr. Crop Sci. J. 4, 479–495.
Peter, J. G. (1995). Reversible jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika 82, 711–732. doi: 10.1093/biomet/82.4.711
Pita, J. S., Fondong, V. N., Sangar,é, A., Otim-Nape, G. W., Ogwal, S., Fauquet, C. M., et al. (2001). Recombination, pseudorecombination and synergism of geminiviruses are determinant keys to the epidemic of severe cassava mosaic disease in Uganda. J. Gen. Virol. 82, 655–665. doi: 10.1099/0022-1317-82-3-655
Ramkat, R., Calari, A., Maghuly, F., and Laimer, M. (2011). Occurrence of African cassava mosaic virus (ACMV) and East African cassava mosaic virus-Uganda (EACMV-UG) in Jatropha curcas. BMC Proc. 5, 93. doi: 10.1186/1753-6561-5-S7-P93
Rey, C., and Vanderschuren, H. (2017). Cassava mosaic and brown streak diseases: current perspectives and beyond. Annu. Rev. Virol. 4, 429–452. doi: 10.1146/annurev-virology-101416-041913
Rey, M. E. C., Ndunguru, J., Berrie, L. C., Paximadis, M., Berry, S., Cossa, N., et al. (2012). Diversity of dicotyledenous-infecting geminiviruses and their associated DNA molecules in Southern Africa, including the South-west Indian Ocean Islands. Viruses. 4, 1753–1791. doi: 10.3390/v4091753
Scholthof, K.-. B. G., Adkins, S., Czosnek, H., Palukaitis, P., Jacquot, E., Hohn, T., et al. (2011). Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 1, 938–954. doi: 10.1111/j.1364-3703.2011.00752.x
Seal, S. E., VandenBosch, F., and Jeger, M. J. (2006). Factors influencing begomovirus evolution and their increasing global significance: Implications for sustainable control. CRC Crit Rev Plant Sci. 25, 23–46. doi: 10.1080/07352680500365257
Sseruwagi, P., Legg, J. P., Maruthi, M. N., Colvin, J., Rey, M. E. C., Brown, J. K., et al. (2005). Genetic diversity of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) populations and presence of the B biotype and a non-B biotype that can induce silver-leaf symptoms in squash, in Uganda. Ann. Appl. Biol. 147, 253–265. doi: 10.1111/j.1744-7348.2005.00026.x
Sseruwagi, P., Maruthi, M. N., Colvin, J., Rey, M. E. C., Brown, J. K., Legg, J. P., et al. (2006). Colonization of non-cassava plant species by cassava whiteflies (Bemisia tabaci) in Uganda. Entomol. Exp. Appl. 119, 145–153. doi: 10.1111/j.1570-7458.2006.00402.x
Tajebe, L. S., Boni, S. B., Guastella, D., Cavalieri, V., Lund, O. S., Rugumamu, C. P., et al. (2015). Abundance, diversity and geographic distribution of cassava mosaic disease pandemic-associated Bemisia tabaci in Tanzania. J. Appl. Entomol. 139, 627–637. doi: 10.1111/jen.12197
Tiendrébéogo, F., Lefeuvre, P., Hoareau, M., Harimalala, M. A., De Bruyn, A., Villemot, J., et al. (2012). Evolution of African cassava mosaic virus by recombination between bipartite and monopartite begomoviruses. Virol. J. 9, 67. doi: 10.1186/1743-422X-9-67
Tocko-Marabena, B. K., Silla, S., Simiand, C., Zinga, I., Legg, J., Reynaud, B., et al. (2017). Genetic diversity of Bemisia tabaci species colonizing cassava in Central African Republic characterized by analysis of cytochrome c oxidase subunit I. PLoS ONE 12, e0182749. doi: 10.1371/journal.pone.0182749
Tomlinson, K. R., Bailey, A. M., Alicai, T., Seal, S., and Foster, G. D. (2017). Cassava brown streak disease: historical timeline, current knowledge and future prospects. Mol. Plant Pathol. 19, 1282–1294. doi: 10.1111/mpp.12613
USAID (2017a). Climate change risk in Karamoja, Uganda: Climate screening for food security. Available online at: https://www.usaid.gov/sites/default/files/documents/1866/170130_Karamoja_Food_Security_Climate_Screening.pdf (accessed on May 15, 2020).
USAID (2017b). Climate risks in food for peace geographies: Karamoja region, Uganda. Available online at: https://www.climatelinks.org/resources/climate-change-risk-profile-climate-risk-screening-food-security-karamoja-region-uganda (accessed on June 12, 2019).
Vyskočilová, S., Seal, S., and Colvin, J. (2019). Relative polyphagy of “Mediterranean” cryptic Bemisia tabaci whitefly species and global pest status implications. J Pest Sci. 92, 1071–1088. doi: 10.1007/s10340-019-01113-9
Vyskočilová, S., Tay, W. T., van Brunschot, S., Seal, S., and Colvin, J. (2018). An integrative approach to discovering cryptic species within the Bemisia tabaci whitefly species complex. Sci. Rep. 8, 10886. doi: 10.1038/s41598-018-29305-w
Werle, E., Schneider, C., Renner, M., Vö1ker, M., and Fiehn, W (1994). Convenient single-step, one tube purification of PCR products for direct sequencing. Nucleic Acids Res. 22, 4354–4355. doi: 10.1093/nar/22.20.4354
White, J. A., Kelly, S. E., Perlman, S. J., and Hunter, M. S. (2009). Cytoplasmic incompatibility in the parasitic wasp Encarsia inaron: Disentangling the roles of Cardinium and Wolbachia symbionts. Heredity 102, 483–489. doi: 10.1038/hdy.2009.5
Winter, S., Koerbler, M., Stein, B., Pietruszka, A., Paape, M., Butgereitt, A., et al. (2010). Analysis of cassava brown streak viruses reveals the presence of distinct virus species causing cassava brown streak disease in East Africa. J. Gen. Virol. 91, 1365–1372. doi: 10.1099/vir.0.014688-0
Wosula, E. N., Chen, W., Fei, Z., and Legg, J. P. (2017). Unravelling the genetic diversity among cassava Bemisia tabaci whiteflies using NextRAD sequencing. Genome Biol. E9, 2958–2973. doi: 10.1093/gbe/evx219
Zerbini, F. M., Briddon, R. W., Idris, A., Martin, D. P., Moriones, E., Navas-Castillo, J., et al. (2017). ICTV Virus Taxonomy Profile: Geminiviridae. J. Gen. Virol. 98, 131–133. doi: 10.1099/jgv.0.000738
Keywords: agro-ecological regions, Bemisia tabaci, host-plants, whitefly species, SSA3, Uganda
Citation: Namuddu A, Seal S, van Brunschot S, Malka O, Kabaalu R, Morin S, Omongo C and Colvin J (2023) Distribution of Bemisia tabaci in different agro-ecological regions in Uganda and the threat of vector-borne pandemics into new cassava growing areas. Front. Sustain. Food Syst. 7:1068109. doi: 10.3389/fsufs.2023.1068109
Received: 12 October 2022; Accepted: 16 January 2023;
Published: 03 February 2023.
Edited by:
Fidèle Tiendrebeogo, Institut de l'Environnement et de la Recherche Agricole (INERA), Burkina FasoReviewed by:
Everlyne Nafula Wosula, International Institute of Tropical Agriculture, TanzaniaFrancis Onyilo, Muni University, Uganda
Habibu Mugerwa, University of Georgia, Griffin Campus, United States
Copyright © 2023 Namuddu, Seal, van Brunschot, Malka, Kabaalu, Morin, Omongo and Colvin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Annet Namuddu,  anamudduojara@gmail.com
anamudduojara@gmail.com
 Annet Namuddu
Annet Namuddu Susan Seal
Susan Seal Sharon van Brunschot
Sharon van Brunschot Osnat Malka3
Osnat Malka3  Richard Kabaalu
Richard Kabaalu Shai Morin
Shai Morin Christopher Omongo
Christopher Omongo John Colvin
John Colvin